Understanding stem cell states • Capturing naive human pluripotency • Defining naive human pluripotency • Epigenetic stability of naive stem cells • Trophoblast stem cells • Stem-cell-derived trophoblast organoids • Modeling human embryogenesis using 3D-cultured blastoids • Future directions
Understanding different states of pluripotency
Embryonic stem cells (ESCs) have the ability to self-renew indefinitely while maintaining the capacity to differentiate into all cell types found in the body. Due to these unique properties, ESCs have become a versatile tool in wide-ranging biomedical applications, from disease modeling to toxicology testing to clinical trials. In addition, the discovery of induced pluripotent stem cells (iPSCs) provides new possibilities to model complex genetic disorders and a source of autologous cells for transplantation. However, major challenges must be overcome before human ESCs and iPSCs can be used in a realistic way in regenerative medicine. The main challenge is that current human ESCs and iPSCs do not resemble the ground state “naive” pluripotent cells found in the blastocyst, but instead are more similar to “primed” precursors that arise after the embryo has implanted. The naive state is signified by an unrestricted developmental potential, whereas the primed state displays repressive chromatin features and lineage priming (Figure 1).
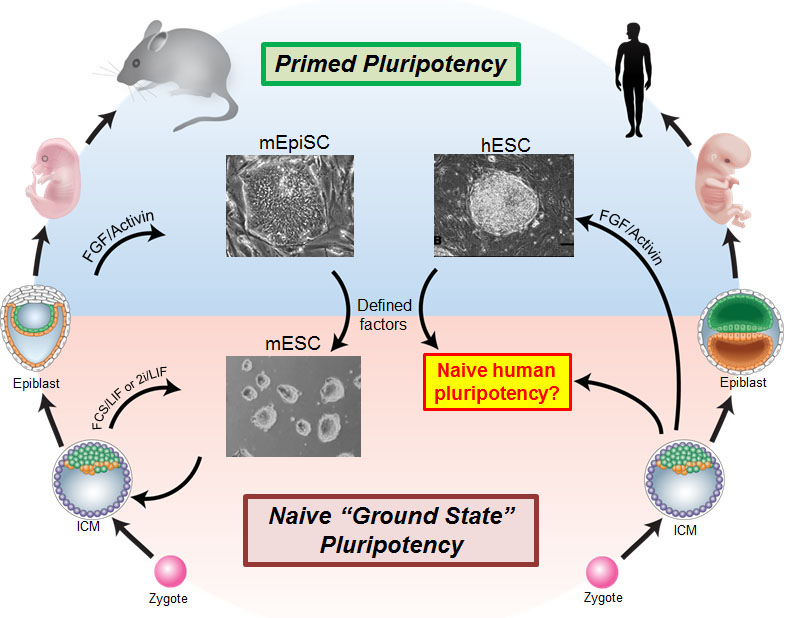
Figure 1. Overview of pluripotent stem cell states in mouse and human. Two distinct pluripotent stem cell states have been stably isolated from mouse embryos: embryonic stem cells (ESCs) derived from the pre-implantation blastocyst are considered to be in a “naive” pluripotent state, whereas epiblast stem cells (EpiSCs) derived from the post-implantation epiblast are in a “primed” pluripotent state. The naive state has an unbiased developmental potential, while the primed state displays lineage priming and repressive chromatin. Human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) most closely correspond to primate post-implantation embryos. There has been significant interest in isolating human ESCs and iPSCs in a naive state that faithfully resembles the human pre-implantation blastocyst. Figure adapted from Dong, Fischer and Theunissen, Exp. Cell Res., 2019.
While naive stem cells can be derived in rodents, their isolation has long remained elusive in the human system. The discovery of naive human pluripotent stem cells (hPSCs) has broad implications for biomedical research. First, naive hPSCs may offer an enhanced starting point for differentiation into disease-relevant cell types, overcoming the heterogeneity frequently observed in current hPSCs. Second, the isolation of naive hPSCs may provide a cell culture system to study epigenetic mechanisms of human pre-implantation development that cannot be investigated in primed cells. Such studies are essential to help understand the high percentage of unexplained pregnancy loss. Third, naive induction may correct the erosion of dosage compensation prevalent in female hPSC lines, enabling faithful in vitro modeling of X-linked diseases, such as mental retardation and autism spectrum disorders. Fourth, the injection of naive hPSCs into the blastocyst of an animal host may allow the generation of interspecies chimeras, providing a novel paradigm to study functional cells derived from patient-specific iPSCs in vivo.
Capturing naive human pluripotency
In order to isolate candidate naive hESCs, we first established a naive-specific fluorescent reporter system. Experiments in mice have shown that the expression of OCT4, a master regulator of pluripotency, is controlled by distinct enhancer elements in the naive and primed pluripotent states. By generating hESCs carrying a deletion of the proximal enhancer in an OCT4-GFP reporter allele, activation of the naive-specific distal enhancer can be monitored in real time. Primed mouse EpiSCs can be reprogrammed to naive pluripotency by overexpression of defined transcription factors and switching to naive-specific culture conditions. Using a similar strategy, we found that inducible expression of KLF2 and NANOG could induce a putative naive state in hESCs. By screening a small molecule library, we identified a combination of five kinase inhibitors that could maintain naive reporter activity in the absence of transgene expression. These conditions also enabled the isolation of naive hESCs directly from primed cells, blastocysts or somatic cells. Naive hESCs are characterized by the upregulation of naive-specific transcription factors and display a reduction in repressive chromatin features (Figure 2).
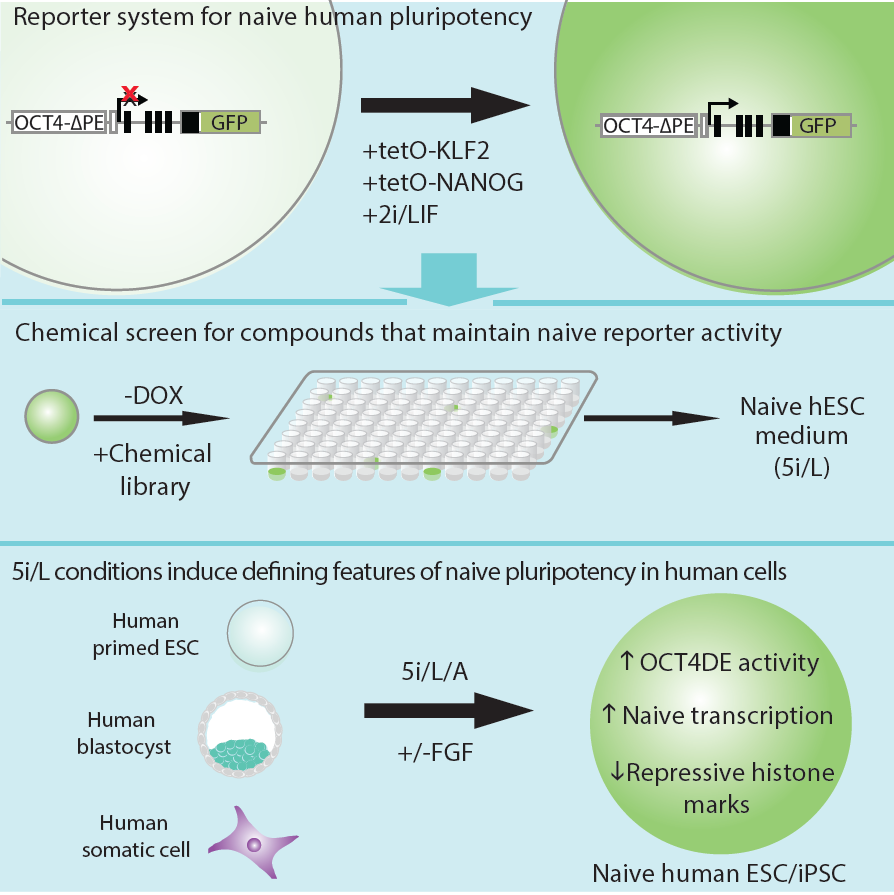
Figure 2. Strategy for isolating naive human pluripotent stem cells. An OCT4-GFP reporter allele containing a deletion of the proximal enhancer was activated by Doxycycline (DOX)-inducible expression of KLF2 and NANOG and switching to 2i/LIF conditions. Screening a small molecule library identified a combination of five kinase inhibitors (5i) that could maintain OCT4-ΔPE-GFP reporter activity upon DOX withdrawal. When combined with recombinant FGF and Activin, these conditions enable the direct transgene-free derivation of naive hESCs and iPSCs from primed cells, explanted blastocysts or somatic cells. Figure adapted from Theunissen et al., Cell Stem Cell, 2014.
The correspondence between stem cells and early human embryos
Studies in recent years have revealed substantial differences in early embryogenesis between mouse and human. These differences include the lack of diapause in human, divergent responses of mouse and human embryos to FGF/Erk inhibition, and differences in the spatiotemporal expression of key developmental regulators and mechanisms of dosage compensation. Based on these observations, an accurate interpretation of human stem cell states must rely on direct comparison with human pluripotent cells in vivo, rather than extrapolation from rodent models. We therefore compared naive and primed hESCs to early human embryos using stringent molecular criteria (Figure 3).
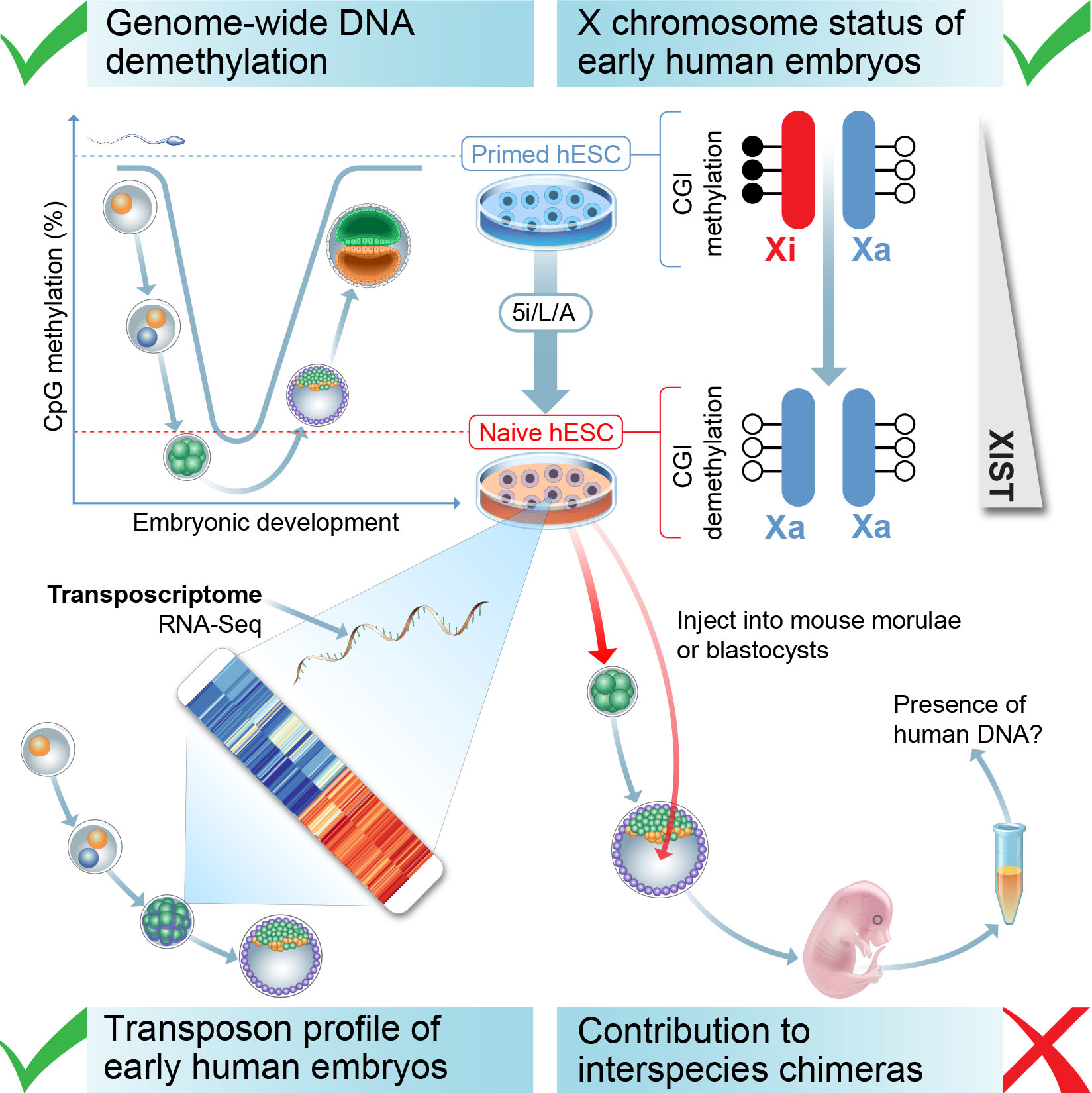
Figure 3. Criteria for defining naive human pluripotency. We have compared candidate naive human PSCs to early embryos using four criteria: (i) The correspondence between transposon expression in naive human ESCs and early stages of human embryogenesis; (ii) The levels of global DNA methylation; (iii) The status of the X chromosome in female cells; (iv) The ability of naive human ESCs to incorporate into mouse morula or blastocyst-stage embryos and contribute to mid-gestation interspecies chimeras. Figure from Theunissen et al., Cell Stem Cell., 2016.
We examined the expression of transposable elements in human pluripotent stem cells and early embryonic stages. Because transposable elements are highly abundant (our genome contains approximately 4 million individual transposable integrants compared to 25,000 genes), the expression of transposons provides a highly sensitive measure of cellular identity. Naive cells in 5i/L/A acquire a transposon transcription signature that closely resembles human morula and blastocyst stage embryos. In addition, base-resolution bisulfite sequencing revealed that naive induction is accompanied by genome-wide depletion in DNA methylation, which is reversible upon differentiation except at imprinted regions. Finally, induction of naive human pluripotency in female cells involves the reactivation of an inactive X chromosome. These results demonstrate that naive human pluripotent stem cells share several defining features with pre-implantation embryos.
Enhancing the epigenetic stability of naive stem cells
An important limitation of current protocols for inducing naive human pluripotency is the erosion of parent-specific epigenetic marks (so-called “imprints”). Imprints are protected during the genome-wide wave of demethylation in the pre-implantation embryo and ensure monoallelic expression of hundreds of genes during human development. Consequently, demethylation of imprints under current conditions for inducing naive human pluripotency creates an epigenetic imbalance in naive stem cells and their derivatives. To track the dynamics of imprint erasure during naive resetting in real time, we established a dual-colored fluorescent reporter at both alleles of the imprinted SNRPN locus. During primed-to-naive resetting, SNRPN expression becomes biallelic in most naive cells, and biallelic SNRPN expression is irreversible upon re-priming. We utilized this live-cell reporter to evaluate chemical and genetic strategies to minimize imprint erasure (Figure 4). Decreasing the level of MEK/ERK inhibition or overexpressing the KRAB zinc-finger protein ZFP57 protected a subset of imprints during naive resetting. Combining these two strategies protected imprint levels to a further extent than either strategy alone. This study offers an experimental tool to track and enhance imprint stability during transitions between human pluripotent states in vitro.
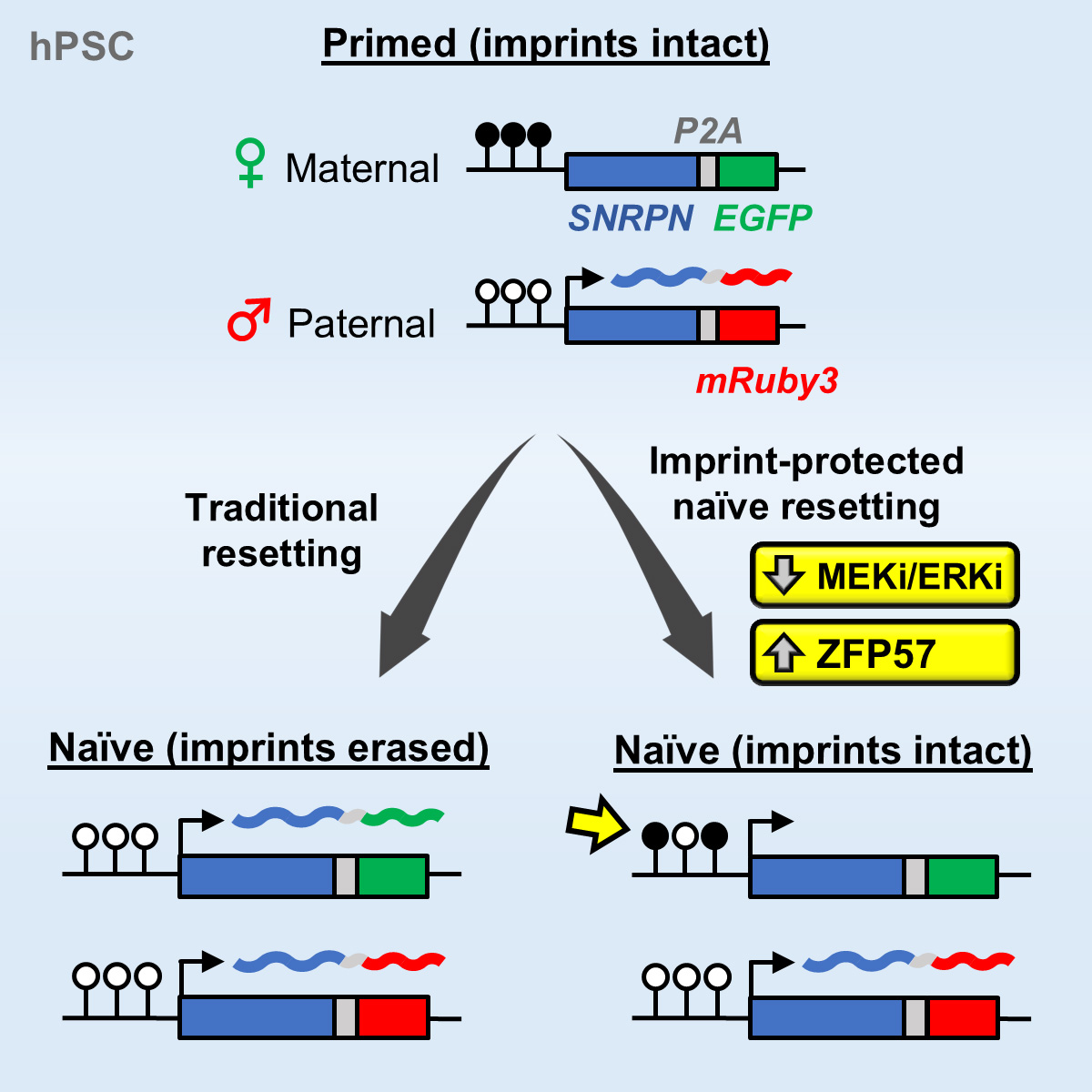
Figure 4. Tracking and mitigating imprint erasure during induction of naive human pluripotency. Current conditions for inducing naive human pluripotency result in the irreversible loss of parent-specific epigenetic marks (“imprints”). We have established a dual-colored fluorescent reporter at both alleles of the SNRPN locus to track imprint erasure during naive resetting. Decreasing the level of MEK/ERK inhibition or overexpressing the KRAB zinc-finger protein ZFP57 protects a subset of imprints during naive resetting. Figure adapted from Fischer et al., Stem Cell Reports, 2025.
Derivation of human trophoblast stem cells (hTSCs) from naive human pluripotent stem cells (hPSCs)
The placenta is a critical organ system that mediates the exchange of nutrients, gases and waste products between the mother and the developing fetus. Placental abnormalities in the first trimester are associated with pregnancy complications such as preeclampsia, miscarriage and fetal growth restriction. However, the placenta is also considered the least understood human organ since access to first-trimester placental tissue is scarce and animal models inadequately recapitulate human placental development. Trophoblast constitutes the predominant epithelial cell type in the placenta and originates from the outer layer of cells in the blastocyst, called the trophectoderm. Given the ethical and legal constraints of studying placental development in vivo, there has been significant interest in establishing reliable in vitro model systems of human trophoblast specification and differentiation.
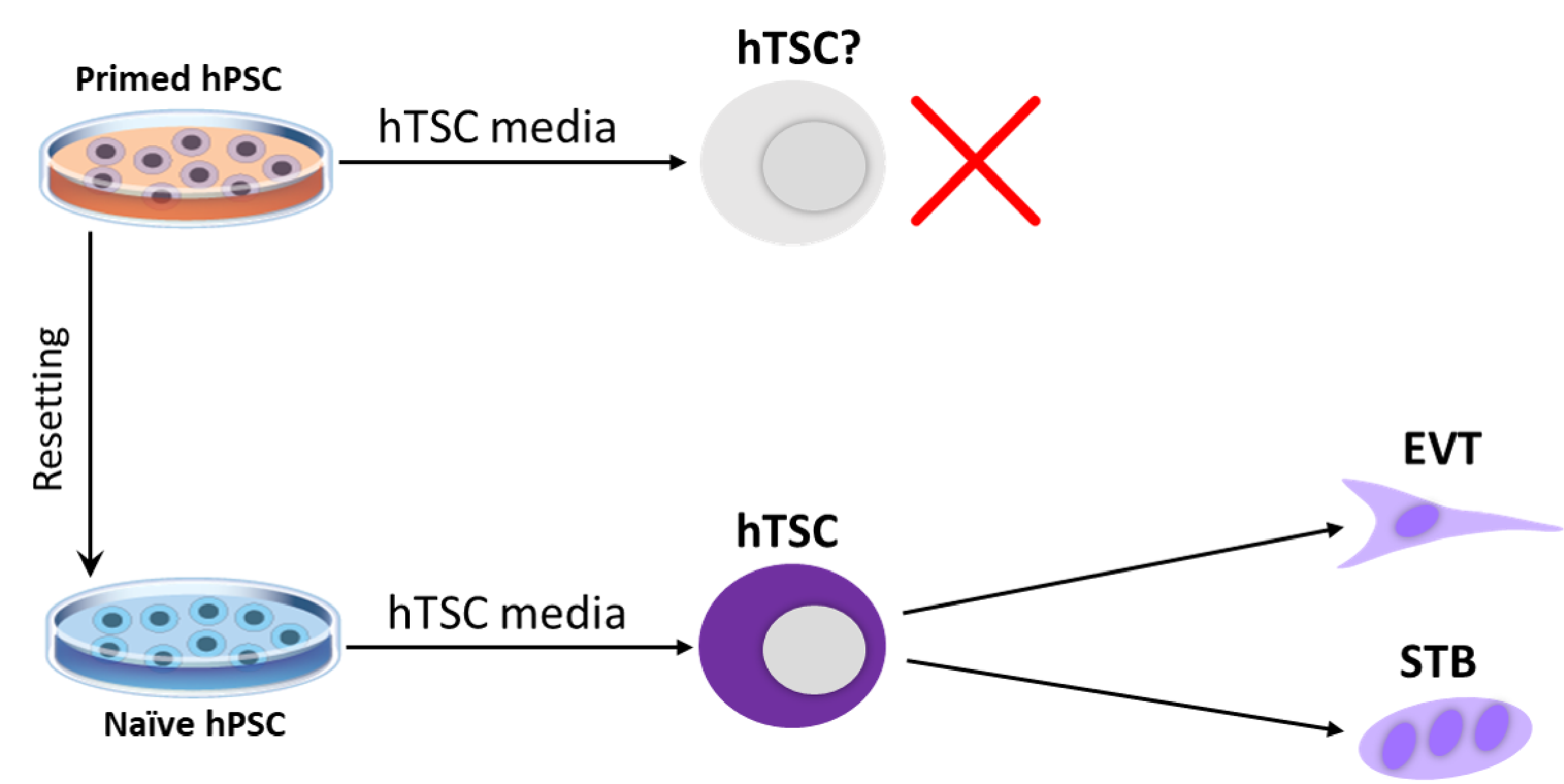
Figure 5. Derivation of human trophoblast stem cells (hTSCs) from naive human pluripotent stem cells (hPSCs). Isogenic naive and primed hPSCs were treated with hTSC media. While primed hPSCs acquired a neural morphology, naive hPSCs differentiated into cells that closely resembled primary hTSCs based on morphological, transcriptional and epigenetic criteria. Furthermore, naive hPSC-derived hTSCs were capable of undergoing cell-type-specific differentiation into specialized trophoblast cell types. Figure adapted from Dong et al., eLife, 2020.
Since naive hPSCs share gene expression and chromatin accessibility signatures with trophoblast cells, we examined their ability to differentiate into this extra-embryonic lineage. We seeded isogenic naive and primed hPSCs in a culture cocktail that promotes the self-renewal of human trophoblast stem cells (hTSCs) (Figure 5). Naive, but not primed, hPSCs acquired typical hTSC-like morphology within several passages. We proceeded to further characterize the naive hPSC-derived hTSCs (referred to as “naive hTSCs”), and found that they express key trophoblast markers at both the mRNA and protein level. We then subjected these naive hTSCs to conditions that promote terminal differentiation towards the extravillous trophoblast (EVT) and syncytiotrophoblast (STB) lineages. The resulting cells satisfy both morphological and molecular criteria of EVT and STB, respectively. Finally, we found that naive hTSCs closely resemble primary hTSCs based on their transcriptome and chromatin accessibility landscape and correspond to a post-implantation TE identity. This work establishes a novel cellular model system to elucidate mechanisms of human trophoblast specification, which is historically difficult to study due to restrictions on the manipulation of human embryos. As a starting point for defining candidate genetic regulators, we performed a genome-wide CRISPR/Cas9 screen to identify essential and growth-restricting genes in hTSCs (Dong et al., Nature Communications, 2022). The candidates identified in this screen provide a framework for elucidating conserved and species-specific regulators of trophoblast development.
Stem-cell-derived trophoblast organoids (SC-TOs): an accessible 3D model system of human placental developmental and disease
In addition to establishing 2D hTSCs from naive hPSCs, we explored whether naïve hPSCs may provide an accessible source of 3D trophoblast organoids that comprise both trophoblast progenitor and differentiated cell types. While such organoids were previously derived from first-trimester placental tissues, access to these tissues is limited due to ethical and regulatory constraints. Indeed, when transferred to Matrigel in appropriate media, naïve-hPSC-derived hTSCs readily self-organize into 3D organoids that exhibit an outer layer of progenitor cells and an inner syncytial compartment (Figure 6a). Single cell transcriptome profiling on these stem-cell-derived trophoblast organoids (SC-TOs) generated from naïve and primary hTSCs revealed a highly concordant cellular distribution of progenitor and specialized trophoblast states (Figure 6b). Transcriptome profiling also revealed a close alignment to trophoblast subtypes found in the early human post-implantation embryo. We further showed that SC-TOs generated from female naïve hPSCs undergo X chromosome inactivation (XCI) and recapitulate patchy XCI patterns seen in human placental tissues (Figure 6c).
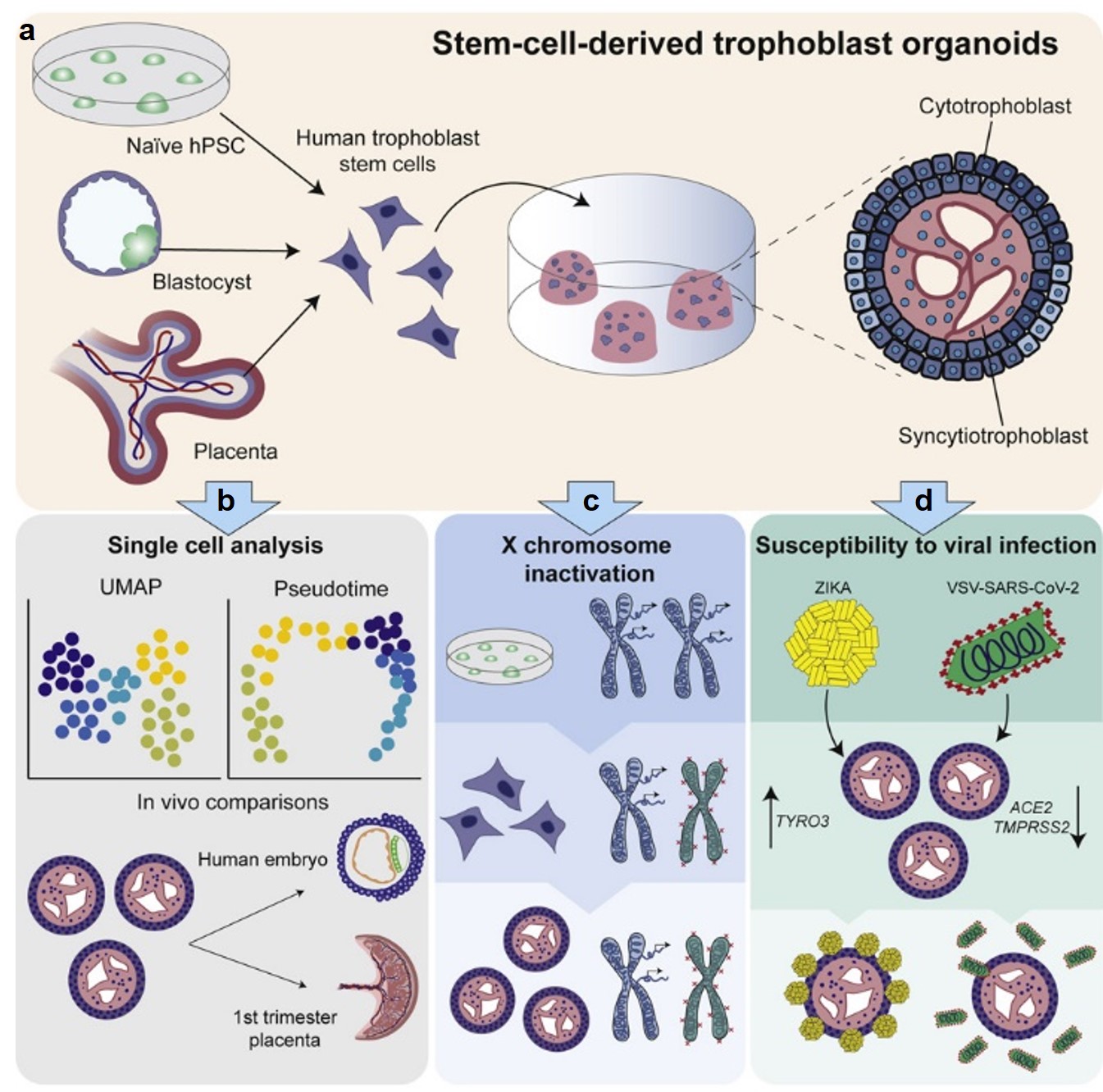
Figure 6. Generation of stem-cell-derived trophoblast organoids (SC-TOs). (a) Human trophoblast stem cells (hTSCs) isolated from naïve hPSCs or primary tissues can self-organize into 3D trophoblast organoids with an inner syncytial compartment and outer shell of cytotrophoblast (CTB) progenitors. (b) Single cell transcriptome studies revealed close alignment between stem-cell-derived trophoblast organoids (SC-TOs) and trophoblast cell types in the early post-implantation embryo (embryonic day 7-14). (c) Female naïve hPSCs undergo X chromosome inactivation upon differentiation into hTSCs and organoid formation results in the clonal expansion of maternal or paternal XCI patterns, as seen in the human placenta. (d) SC-TOs display selective vulnerability to SARS-CoV-2 and Zika viruses, which correlates with relative expression of viral entry factors. This graphical abstract summarizes our recently published findings (Karvas et al., Cell Stem Cell, 2022).
We investigated whether SC-TOs may be useful to model placental susceptibility to emerging viral infections. Whereas SC-TOs were readily infected by Zika virus, they demonstrated only limited susceptibility to SARS-CoV-2, specifically within a subset of syncytiotrophoblast cells (Figure 6d). This selective vulnerability correlates with differential expression levels of the entry factors for these two viruses. These studies demonstrate that naïve hPSCs provide an accessible source of 3D trophoblast organoids, which can be used to model the developing placenta and its vulnerability to emerging pathogens. Future efforts in our lab will exploit this 3D organoid model to investigate placental-endometrial crosstalk and the genetic basis of human placental development.
Building a continuous and integrated model of human embryogenesis using naive hPSCs
Our work and that of others demonstrated that naive hPSCs have a remarkable capacity for extraembryonic differentiation, readily differentiating into trophoblast and primitive endoderm in vitro. This raised the exciting possibility that naive hPSCs may have the capacity to self-organize into a complete model of the human embryo that comprises both embryonic and extraembryonic lineages. Indeed, in a transformative advance, several groups reported that naive hPSCs can form blastocyst-like structures (also known as “blastoids”) that model the human pre-implantation embryo. However, the extent to which blastoids can recapitulate defining features of post-implantation development remained unexplored. We have optimized the conditions for blastoid generation from 5i/L/A naïve hPSCs and investigated their capacity for extended culture on thick 3D extracellular matrices, which better mimic the physical environment of the human endometrium compared to flat surfaces. We developed an experimental methodology that supports human blastoid culture for up to day 21, including the formation of complex embryonic and placental structures (Figure 7). By performing a detailed single cell transcriptome analysis at three distinct time points (D7, D14, and D21), we benchmarked our model system to human embryos at pre-implantation, early post-implantation, and early gastrulation stages.
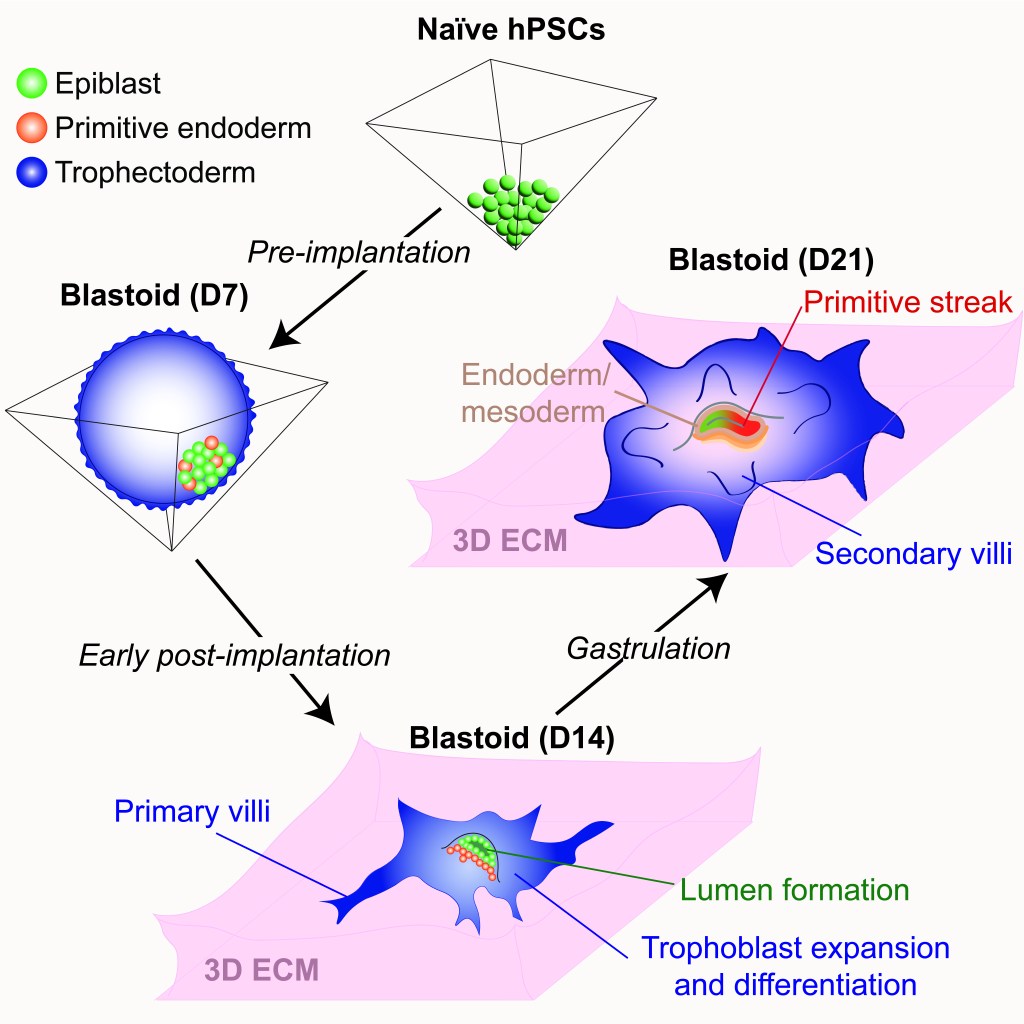
Figure 7. 3D-cultured blastoids model human embryogenesis from pre-implantation to early gastrulation stages. By promoting the self-organization of naive hPSCs into blastoids and allowing their extended culture on 3D extracellular matrices, we have established an integrated model system of early human embryogenesis spanning pre-implantation and early post-implantation development up to the formation of a primitive streak-like structure and the induction of definitive endoderm and mesoderm lineages. This model system enables continuous in vitro monitoring of human embryonic development across the implantation window, while the trophoblast compartment expands and differentiates into specialized placental cell types (Karvas et al., Cell Stem Cell, 2023).
3D-cultured human blastoids display several molecular and morphogenetic hallmarks of early post-implantation development, including lumenogenesis of the epiblast compartment, rapid expansion and diversification of trophoblast lineages, and robust invasion of extravillous trophoblast cells by D14. Extended blastoid culture resulted in the formation of a primitive streak-like structure, as evidenced by the localized activation of TBXT (Brachyury) by D18. Blastoids maintained until D21 acquired a single cell transcriptome profile that closely resembled that of a gastrulating human embryo analyzed at Carnegie Stage 7. This included the emergence of blastoid primordial germ cells, definitive endoderm, and various mesodermal lineages, and a diverse array of extraembryonic cell types, including blastoid amnion, cytotrophoblast, extravillous trophoblast, extraembryonic mesoderm, syncytiotrophoblast, and yolk sac endoderm. Thus, the 3D-cultured human blastoids described herein model embryonic and extraembryonic development from pre-implantation to early gastrulation stages, offering a continuous and integrated in vitro model system of early embryogenesis. We propose that this model system will allow investigation of the genetic and epigenetic mechanisms regulating both implantation and gastrulation, two critical bottlenecks for successful pregnancy that involve extensive interactions between embryonic and extraembryonic tissues.
Future directions
The above studies indicate that naive hPSCs offer a window into mechanisms of human pre-implantation development that are difficult to model in conventional hPSCs, such as the regulation of X chromosome reactivation and inactivation, the activity of early embryonic transposons, the mechanisms of trophoblast specification, and the spontaneous generation of integrated human embryo models. However, current naive hPSCs do not perfectly resemble pluripotent cells in the human blastocyst, as exemplified by their global loss of imprinting and non-random XCI during embryonic differentiation. Thus, it remains to be determined whether strategies can be devised to capture naive hPSCs that are as robust as their mouse counterparts, which may facilitate wide-ranging applications in regenerative medicine. Additionally, while 3D-cultured blastoids harbor most embryonic and extraembryonic cell types seen in human embryos at corresponding stages of development, some cell types emerged with delayed kinetics, such as the amnion and yolk sac lineages. Therefore, continued refinement of the conditions for extended blastoid culture is required to faithfully reconstruct the complex interactions between embryonic and extraembryonic lineages in vitro.
Our current research is focused on three broad questions: (i) What are the signaling requirements for naive pluripotency, and how can we manipulate these pathways to create an optimal culture environment for deriving and maintaining human ESCs and iPSCs? (ii) What are the major transcriptional and epigenetic mechanisms underpinning the self-renewal and lineage commitment of pluripotent and trophoblast stem cells? (iii) Can we leverage recently established stem-cell-based models of the human embryo and placenta to obtain mechanistic insights into early human development with therapeutic relevance for advancing maternal and fetal health?


